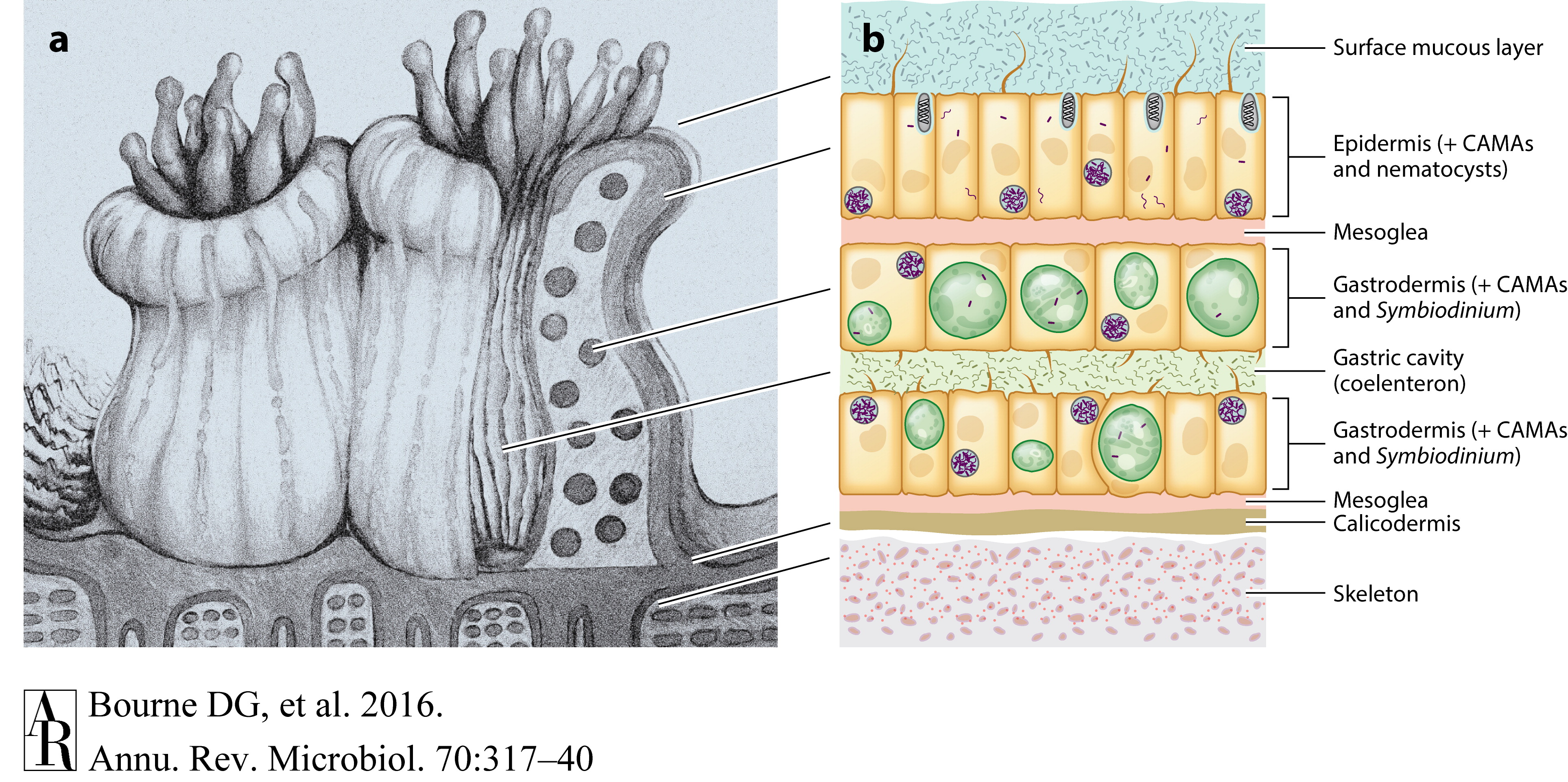
Daily Lab Log
04-MAY-2023. W
- received Qubit DNA Broad Range (BR) Assay Kit!
- Gotta get P2 DNA/RNA free tips!
- ran qubit on DNA & RNA, got great results!
25-APR-2023. T
- sorted pipettes with Callum
- submitted Bioinformatics Assignment
- ordered Qubit DNA BR assay kit
- ordered film sealing plate roller
24-APR-2023. M
-
sent draft letter for NAH experiential learning fund to JPG on Monday April 24th
-
emailed Mr.DNA about 20 16S samples
-
read up on NanoDrop vs. Qubit
-
reached out to Sam White about Ubuntu/Windows dual-boot computer in lab 236, which is connected to the Thermo NanoDrop 2000c Sam shared:
FYI, if you’re solely using the NanoDrop for quantification (i.e. not worried about 260/280), then I’d highly recommend just relying on the Qubit, with both DNA and RNA. The reason for this is that the NanoDrop will almost always over-“calculate” your sample concentrations (both RNA and DNA). The reason for this is that the NanoDrop is a spectrophotometer and measures light absorbance. With almost all DNA/RNA isolations, there will be residual DNA or RNA in your sample. The NanoDrop cannot distinguish between the two molecules, because they’re both made up of the same material; nucleotides. Nucleotides, regardless of their of their origin will absorb light at an average wavelength of 260nm.Since the Qubit assays use molecule-specific dyes, the dye will only bind to DNA or only to RNA. The dyes will not bind to any carryover of other types of molecules in the sample, so the accuracy (and sensitivity) is much, much higher than the NanoDrop.
21-APR-2023. F
- met with JPG April 1on1
- talked about travel from Sept. 12 - Oct 9th, missing first 2 weeks of Autumn ‘23 quarter, focusing that quarter on writing & analysis
- GOAL: have all sequences (16S & Transcriptomes) by September 1st 2023
- Initial 20 samples extracted & sent for sequencing before FHL
- 2nd committee meeting to be held once initial 20 samples are run through 1st analysis for 16S & DGE, get advice on strategy for selecting remaining samples
- Follow-up samples extracted & sent for sequencing in July/August
- Send draft letter for NAH experiential learning fund to JPG on Monday April 24th
- Initial test round 2 samples extracted!!!
- 3-CA1a
- 3-Ea
- Sampes selected from extr1 using a random number generator
- details can be found in the coral-DNA-RNA-lab-extractions repository
20-APR-2023. Th
- downloaded fastq files for ‘fake data’ to continue script for bioinformatics workflow
- Received Mortexer! Whoo!
- Received cryo-gloves
- got 8lbs of dry-ice with NH
- re-organized cryo-vials to wax boxes, with all of the ‘a’ samples in 2 boxes labelled with green lab tape and ‘b’ samples in 2 boxes labelled with blue tape with NH
- setup lab bench for dry run dna/rna extraction with NH
- NH setup mortexer
- submitted 2-year extended warranty for mortexer s/n 22040448 via benchmark scientific in JPG’s name
- placed signage on lab 236 to lint roll and lab coat (because I forgot to and that was embarassing)
- submitted purchase for mortexer attachment head that will homogenize 12 1.5-2mL tubes at a time horizontal orientation (even better because closer to original lysis protocol for Zymo) Sadly, I should have looked for something like this earlier, but I saw it on the mortexer instruction manual when it was delivered and realized that was what I needed
19-APR-2023. W
- continued draft application for ONeill-Quinn
- continued editing extraction protocol
- expected delivery of mortexer by end of day
18-APR-2023. T
- Met with Nora, selected samples for processing using R script to randomize
- Setup workflow in Hyak
- added to extraction protocol
- waiting for mortexer delivery, expected by end of day, but FedEx package delivery delayed
- emailed asking for details about Neptune controller unit (it was returned after being sent away for RMA maintenance request, but nothing appears to have been done to it)
17-APR-2023. M
- added to extraction protocol
- read about remote computing, Hyak Mox
- enabled Hyak account
14-APR-2023. F
- attended Cat Fong chalk talk & SAFS Cafe
- gave tour to Edith
11-APR-2023. T
- NCBI SRA links downloading RNAseq fastq files
- Looking for ‘fake data’ for M. capitata
- DNA, ITS2 to measure symbiont to host cell ratios for ‘Patterns of bleaching and recovery of Montipora capitata in Kaneohe Bay’
hosted at zenodo here https://zenodo.org/record/53231#.ZDWE0HbMKTQ
-RNAseq resources from A. Huffmeyer
- https://ahuffmyer.github.io/ASH_Putnam_Lab_Notebook/E5-Deep-Dive-RNAseq-Count-Matrix-Analysis/
- https://github.com/AHuffmyer/EarlyLifeHistory_Energetics
Here is the spreadsheet of my NCBI accession numbers for my sequences: https://docs.google.com/spreadsheets/d/1qDGGpLFcmoO-fIFOPSUhPcxi4ErXIq2PkQbxvCCzI40/edit?pli=1#gid=457642286 If you look at the Mcapitata Ontogeny Timeseries tab it’ll have all the SRR numbers you will need to download the TagSeq files. Instead of downloading the full data set you could just pick like three lifestages for a test comparison - AH
- DNA, ITS2 to measure symbiont to host cell ratios for ‘Patterns of bleaching and recovery of Montipora capitata in Kaneohe Bay’
hosted at zenodo here https://zenodo.org/record/53231#.ZDWE0HbMKTQ
-RNAseq resources from A. Huffmeyer
10-APR-2023. M
- created script to randomize sample processing in coral-DNA-RNA-lab-extractions repo
- called Zymo and learned that ‘mix’ in protocol step can either mean pipette up and down 10-12 times, or briefly touch to vortex mixer for a pulse
- Zymo tech also shared that it’s best to follow protocol and add Lysis buffer to the sample supernatent in a separate rnase-free tube. If you do it in the filter tube some sample may start to percolate thru the filter before being fully buffered.
- Horizontal Microtube Holder
29-MAR-2023. W
- started template for DNA/RNA data extraction sheet in new repo for extractions
- organized notes for samples in metadata
- installed git on WSL via this tutorial
- finished homework 1 for FISH546
- lab-meeting notes
- purchasing
- no purchases in June!
-
The University of Washington is transitioning from their old DOS-based system to Workday Finance, with a Go Live date of July 5th, 2023. During the transition period, there will be some disruptions to normal operations, and the continuity of research and teaching and minimizing the impact on faculty, staff, and students is the priority. There will be Restrictive Periods during which no one will be able to process purchase orders or obtain reimbursements in ARIBA. Exceptions may be made for urgent or emergency purchases. Travel reimbursements and non-travel-related reimbursements should be submitted before June 9th, and processing will resume on July 5th. Blanket Purchase Orders that are expected to be active past June 30th, 2023, will not be automatically transferred to Workday, and all continuing BPOs must be manually reentered by the College of the Environment. Navigating the transition will require patience and close partnerships between research and administrative groups
- space survey
- % of time spent in spaces from July ‘22 - March ‘23
- 2% 242
- 8% 236
- 30% HIMB
- 60% 264, 264B
- undergrad Nora Hessen 2hrs per week in 264, 264B since January ‘23
- purchasing
28-MAR-2023. T
- read papers:
-
called Zymo tech Christian Camacho ccamacho@zymoresearch.com to get advice on working with coral samples and the Zymo DNA/RNA miniprep kit:
- bead bash for 40 minutes for microbes on mortexer vortex-mixer! This is still tested to result in good RNA, though there may be a trade-off between bead-bashing for lysis and RNA interity… see Zymo optimized lysis protocol
- benchmark mortexer is not a tested method for the kit.. but it’s closest to the vortex genie approved method for lysis
- mortar & pestle with liquid nitrogen, then add powder to bead bashing tube with RNA shield
- lyophilized (freeze-dried) proteinase K is stable at room temp, it should be stored in the -20 freezer only after resuspending in the buffer
- the DNAse 1 is similarly a lyophilized powder, but a very small amount, such that the vial looks empty
-
Gathered lab supplies
- mortar & pestles
- alcohol burner for use with denatured ethyl alcohol
- flint spark torch ignitor
- found expired RNA later
- DNAse away
- DEPC water (does that expire?)
- weigh papers (for powdered samples)
NEED
- scoopulas (to scoop powder from mortar to weigh paper)
- denatured ethyl alcohol (disinfect labware between samples)
27-MAR-2023. M
- read papers:
- https://environmentalevidencejournal.biomedcentral.com/articles/10.1186/s13750-023-00298-y
- worked on coral collection report
24-MAR-2023, F
- spring break
####### 23-MAR-2023, Th
- spring break
23-MAR-2023, Th
- spring break
22-MAR-2023, W
- spring break
21-MAR-2023, T
- spring break
- interviewed for WSG student assistant position
- began work on himb-2022 coral collection report
20-MAR-2023, M
- spring break
- installed Sophos anti-virus on Owl
- attended Robert’s Lab meeting
- checked out Qubit buffer + reagents in Roberts lab with Olivia & Zach
- put mortexer, DEPC-treated water, Qubit assay kit, on purchase log
- wrote Qubit RNA Broad Range Protocol post
16-MAR-2023
- finished SMEA/FISH 539 final paper
15-MAR-2023
- out sick
- worked on SMEA/FISH 539 final paper
14-MAR-2023
- out sick
- talked with Hailey Dockery about shadowing RNA extractions
13-MAR-2023
- worked on SMEA/FISH 539 final paper
- updated R & RStudio to latest versions
10-MAR-2023
- finished SMEA/FISH 1page reflection
- worked on SMEA/FISH 539 final paper
- met with GSIS speaker on Zoom
- made hotel reservations for GSIS
09-MAR-2023
- Attended Coastal Ecology candidate chalk talk
- Shadowed Grace for new RNA extraction protocol & Qubit QC for RNA quality
- Met with Emma Strand about M cap DNA/RNA extractions… notes from Zoom chat:
- email Erin Chille eec72@scarletmail.rutgers.edu about 16S extraction kit
- https://emmastrand.github.io/EmmaStrand_Notebook/Kbay-Bleaching-2019-DNA-RNA-Extractions/
- host DNA (looked at DNA methylation), symbiont DNA (ITS2), microbiome DNA (16S not so good), host RNA + symbiont RNA (Illumina Nova Seq RNAseq)
- coral issues- mucus clogging spin-column filter (avoid mucucs, avoid skeleton)
- how big of a fragment do I put in the bead tube? 1/3 bead, 1/3 coral, 1/3 shield – https://emmastrand.github.io/EmmaStrand_Notebook/Kbay-Bleaching-2019-DNA-RNA-Extractions/
- optimize time to vortex bead tube (2mins for m. cap) *vortex machine, holds 8 tubes at once
- https://www.southernlabware.com/mortexertm-vortex-mixer-115v.html?gclid=Cj0KCQiApKagBhC1ARIsAFc7Mc5LH-QjqtDR98g9ZgE9iAOs-y35m7X5rr7pHCqfPGRtTJVPEeQWbLYaAtCpEALw_wcB
- 100uL total DNA material end product from kit
- 12ng of input for ITS2 about 5uL
- aim for 10 - 70 ng/uL DNA quantity (Qubit)
- DNA quality (gel) *important for methylation studies, but may not be for 16S/ITS2
- 100uL total RNA material end product, send 20-30uL for RNAseq
- https://github.com/hputnam/HI_Bleaching_Timeseries/blob/main/Dec-July-2019-analysis/data/Molecular-labwork.xlsx
- sample ID, use a separate extraction ID (in the event of multiple extractions from the same sample!)
07-MAR-2023
- worked on SMEA/FISH 539 final paper
- met to make GSIS budget w/ Juliette
- met with Grace from Robert’s Lab to practice RNA extraction
- participated in SAC interview for ADDEI
06-MAR-2023
- presented final group project in SMEA/FISH 539
- worked on SMEA/FISH 539 final paper
- attended Coastal Ecology faculty candidate seminar
03-MAR-2023
- I swear I’ve done stuff since January / how has winter quarter gone by so fast?!
- submitted PCC Climate Solutions grant funding application this morning
- worked on FSMEA/FISH 539 final project + presentation
02-MAR-2023
- submitted FHL TA application for summer A term marine botany
- helped interview ADDEI candidate
- attended Carter’s Coastal Ecology candidate chalk talk
- met with Steven on Zoom and got permission to use Bioanalyzer
- worked on sequencing quotes, DNA/RNA extraction protocol, reached out to Emma Strand and Grace
31-JAN-2023
- prepared 1st committee meeting blog post
20-JAN-2023
- cleaned PBB
- strategized chapter 1: Low Dose Effects & chapter 2: Multiple Stressors for thesis
- arranged to meet with Miranda to talk about summer kelp gametophyte project
- completed lab safety trainings
19-JAN-2023
- read Ariana Huffmyer’s lab posts on extraction protocols
- read Ainsworth et al 2015 & Brown et al 2021
- MarBio Plant DNA extraction Kit for whole coral holobiont 16S rRNA Bacteria
- Pure Link Plant Total DNA purification systems (Invitrogen) for 16S rRNA Bacteria
- attended JPG lab meeting and gave feedback on Eileen Bates’ pinto abalone presentation for Alaska Science Sympsosium
- designed graphical methods in Canva for pae-temp experiment
18-JAN-2023
- worked with norahess to setup collaboration on ibis repo!
- talking about setting up Aiptasia spawning tanks to run experiments with cnidarian model organism reproduction
17-JAN-2023
- attended Roberts lab meeting, discussed curriculum development for Maritime Highschool
13-JAN-2023
- Read up on ‘Omics’, started a post on definitions and basics of the study of DNA & RNA
- Found whole genome for M. capitata on NCBI
- Helmkampf, M., Bellinger, M. R., Geib, S. M., Sim, S. B., & Takabayashi, M. (2019). Draft Genome of the Rice Coral Montipora capitata Obtained from Linked-Read Sequencing. Genome Biology and Evolution, 11(7), 2045–2054. https://doi.org/10.1093/gbe/evz135
- Williams, A., Pathmanathan, J. S., Stephens, T. G., Su, X., Chiles, E. N., Conetta, D., Putnam, H. M., & Bhattacharya, D. (2021). Multi-omic characterization of the thermal stress phenome in the stony coral Montipora capitata. PeerJ, 9, e12335. https://doi.org/10.7717/peerj.12335
- Shumaker, A., Putnam, H. M., Qiu, H., Price, D. C., Zelzion, E., Harel, A., Wagner, N. E., Gates, R. D., Yoon, H. S., & Bhattacharya, D. (2019). Genome analysis of the rice coral Montipora capitata. Scientific Reports, 9(1), 2571. https://doi.org/10.1038/s41598-019-39274-3
12-JAN-2023
- Explored the use of purple sea urchin and/or Aiptasia for phthalate low-dose effect gametogenesis study (proposal brainstorming) for the IBIS research mentor program
- Created ibis repo to organize project
- Completed lab safety training matrix for myself and NH, identified required and recommended lab safety trainings
- Potential to use rack in PBB near interior door for Aiptasia spawning incubator, setup workflow with dissecting scope and camera all in PBB for IBIS project
- Inventoried equipment we have in lab 236 for molecular work:
- Beckman Coulter Allegra X-30R Centrifuge
- Thermo Scientific NanoDrop 2000c Spectrophotometer
- Fisherbrand Isotemp heat block
- Beckman Coulter Microfuge 16 Centrifuge
- Eppendorf Mastercycler nexus gradient
09-JAN-2023
-
Today I setup my GitHub ‘notebook’! This is the beginning of my exploration of using GitHub to jot down thoughts, record ‘daily bits’, and organize the procedures and methods I’ll use in my thesis research
-
zbengt introduced me to ‘elicit’, a lit review search AI. I searched “What is the role of endocrine signaling in cnidarian development” and found an exciting paper titled “Sea Anemones Responding to Sex Hormones, Oxybenzone, and Benzyl Butyl Phthalate: Transcriptional Profiling and in Silico Modelling Provide Clues to Decipher Endocrine Disruption in Cnidarians” authored by Morgan et al. in 2022
-
I explored another AI, ChatGPT, and gave it the prompt “write me a poem about corals and oysters and mothers” and it returned:
-
Coral reefs, a vibrant sea of life, A home for countless creatures, rife. Oysters, clams, and mussels too, All nestled safe, their shells in view.
But danger lurks, the waters warm, The coral bleaches, it’s colors torn. Mother Nature’s wrath, a sight to see, But we can help, if only we believe.
For mothers, fierce and loving beasts, Will do what it takes, to protect at least. Their young and home, with all their might, They’ll fight and strive, through day and night.
So let us join, in their noble quest, To save the reefs, and all the rest. For oysters, corals, and mothers too, Deserve a chance, to make it through.
-
-
I followed up with a ChatGPT prompt “what are examples of endocrine signaling in cnidarians?” and it returned:
-
Endocrine signaling is the process of communicating information from one cell to another by releasing hormones into the circulatory system or through direct contact. In cnidarians, endocrine signaling can be used for a variety of functions, including the regulation of growth, development, and behavior.
One example of endocrine signaling in cnidarians is the release of growth hormones, which stimulate the growth and development of cells and tissues. For example, in some species of cnidarians, the release of growth hormones is regulated by the action of a hormone called insulin-like growth factor (IGF).
Another example of endocrine signaling in cnidarians is the regulation of reproductive function. In some species of cnidarians, hormones called gonadotropins are released by the endocrine cells of the gonads (sex organs) to stimulate the production and release of gametes (sex cells).
Endocrine signaling is also involved in the regulation of behavior in cnidarians. For example, some species of cnidarians release hormones called neuropeptides that act on the nervous system to regulate behaviors such as feeding, movement, and response to stimuli.
Overall, endocrine signaling plays a vital role in the regulation of various functions in cnidarians, including growth, development, and behavior.
-
**Is this is all true!? I’ll have to follow-up with my own dive into primary literature…
-
Another ChatGPT AI response, when asked the same questions, but prompted to provide references:
-
Examples of endocrine signaling in cnidarians include the release of hormones in response to stimuli such as touch, temperature changes, and changes in light intensity.
For example, the hormone adenylate cyclase-activating polypeptide (ACAP) is released from cnidocytes in response to a variety of stimuli and activates adenylate cyclase, leading to the production of cyclic adenosine monophosphate (cAMP) and the regulation of various physiological processes.
Other signaling molecules that have been identified in cnidarians include cyclic guanosine monophosphate (cGMP), inositol triphosphate (IP3), and Ca2+-dependent signaling pathways.
References:
Levitan, D. R., & Kohn, A. B. (2001). The Endocrine System of Cnidarians. Annual Review of Physiology, 63(1), 319-344.
Schmid, V., & Pfründner, R. (1995). Adenylate cyclase-activating polypeptide in cnidarians: a new class of peptides with signal transducing properties. Regulatory Peptides, 61(2), 149-157.
-