Samples
6C14F (single sample ~10 embryos)
7C14F (single sample ~10 embryos)
6C9F, 7C9F = 67C9F (pooled sample. ~20 embryos)
Summary
No success with RNA quantity as measured by Nanodrop or Broad Range Qubit Assay.
I think I should increase starting sample size, and look into alternative sample prep for delicate embryos (as compared with adult coral fragments).
I can’t be sure that waiting too long during the DNase Treatment didn’t degrade all the RNA, next round be careful not to go over the 15 minute DNase I incubation time.
Extraction Notes
date: 12-AUG-2024
kit: Zymo Quick-DNA/RNA Miniprep Plus Kit
Followed protocol according to my post [M. Capitata Embryo DNA/RNA Extractions Protocol] (https://sarahtanja.github.io/quarto-blog/posts/projects/coral-embryo-leachate/embryo-extractions.html#proteinase-k-digestion) with these changes:
Lysed samples via Bead Bashing in a full 2mL Zymo BeadBashing (0.1 - 0.5mm glass bead) Tube with a total volume of 1mL of DNA/RNA Shield
Transfered 500uL of cleared supernatent to a new nuclease-free tube, making sure not to disturb beads and debris at the bottom of the bead-bashing tube
Conducted Proteinase K digestion as follows:
Get the Proteinase K out of the -20 freezer & set the heat-block to warm up to 55C for 30mins
After bead bashing, tubes are ‘intensely bubbly’, to tamp down bubbles, centrifuge in the mini-centrifuge for 1min
Transfer 500uL of supernatent to a new nuclease-free tube, making sure not to disturb beads and debris at the bottom of the bead-bashing tube2
Add Proteinase K & Buffer
Add the appropriate volume of Pro K buffer and Proteinase K (Proteinase K is stored in the -20 after being reconstituted)
(10:1 ratio of sample:digestion buffer) & (2:1 ratio of digestion buffer:Proteinase K)
For tubes with approximately 500uL of sample add:
50ul pro K buffer 25ul proteinase K Vortex to mix
Incubate in the heat-block for 30mins at 55C
Vortex to mix & centrifuge on max for 2mins to pellet any debris
Transfer 350uL of the cleared supernatent to a new 1.5mL nuclease-free tube, making sure not to disturb any debris at the bottom of the tube
Add 350uL of DNA/RNA Lysis Buffer to the supernatent(1:1) and vortex to mix
I let DNase I treatment incubate at room temperature for 1 hour (instead of 15 minutes)
eluted DNA volume: 30uL in Zymo DNase-RNase Free Water
eluted RNA volume: 30uL in Zymo DNase-RNase Free Water
Nanodrop
RNA
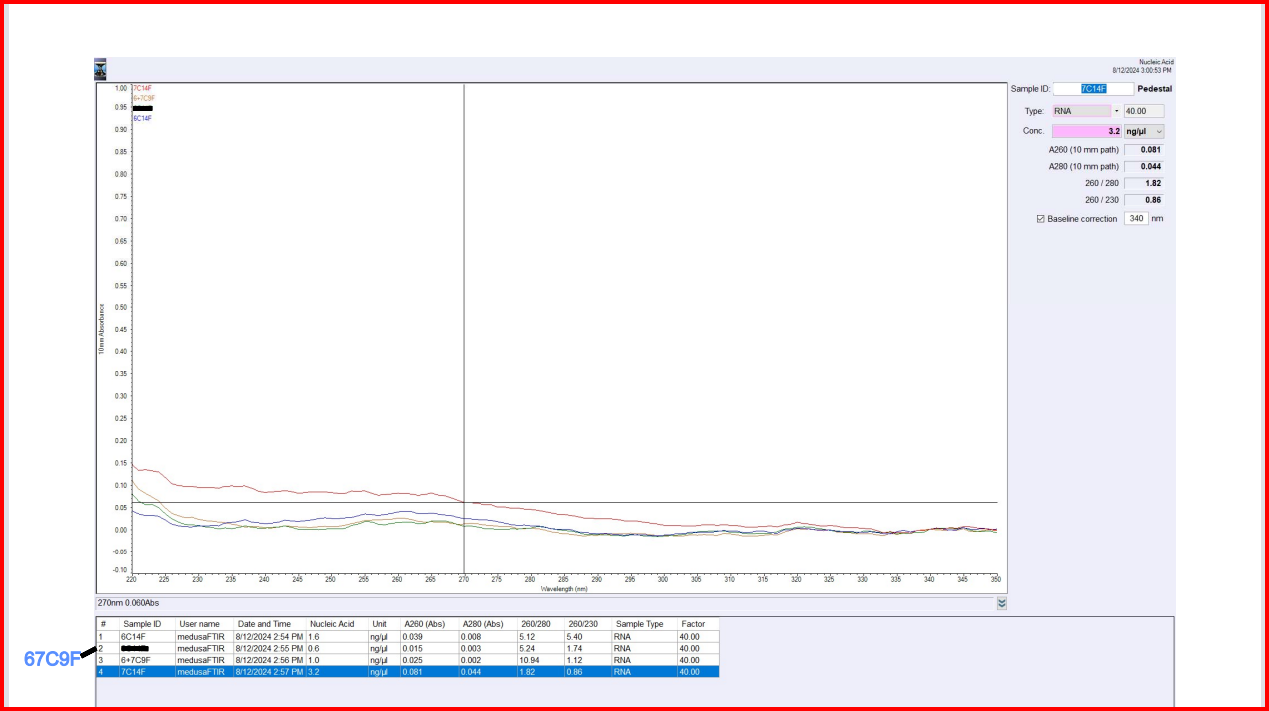
| sample_id | ng/uL | A260 | A280 | 260/280 | 260/230 |
|---|---|---|---|---|---|
| 6C14F | 1.6 | 0.039 | 0.008 | 5.12 | 5.40 |
| 7C14F | 3.2 | 0.081 | 0.044 | 1.82 | 0.86 |
| 67C9F | 1.0 | 0.025 | 0.002 | 10.94 | 1.12 |
Qubit
RNA
Using the Invitrogen Thermo Fischer RNA Broad Range Assay Kit.
Prepared working solution for 3 samples and two standards:
Qubit RNA BR reagent in DMSO = 1uL * 5 = 5uL
Qubit RNA BR Buffer = 199uL * 5 = 995uL
standard 1:
standard 2:
RunID: 2024-08-13_062825
| sample_id | qubit_rna_1 | qubit_rna_2 |
|---|---|---|
| 6C14F | too low | too low |
| 7C14F | too low | too low |
| 67C9F | too low | too low |
Storage Location
RNA samples are stored in -80C ‘Old Friedman’ Sanyo freezer on shelf 1 in coral-embryo-leachate RNA wax freezer boxes with green label tape.
DNA samples (not quantified) are stored in -20C in JPG Lab FSH 236 in shelf 2 in coral-embryo-leachate DNA wax freezer box with yellow label tape.
Protein flow-through (not quantified) are stored in -20C in JPG Lab FSH 236 in shelf 2 in coral-embryo-leachate PROTEIN wax freezer box with blue label tape.