if ("tidyverse" %in% rownames(installed.packages()) == 'FALSE') install.packages('tidyverse')
if ("reshape2" %in% rownames(installed.packages()) == 'FALSE') install.packages('reshape2')
if ("plotly" %in% rownames(installed.packages()) == 'FALSE') install.packages('plotly')
if ("car" %in% rownames(installed.packages()) == 'FALSE') install.packages('car')
if ("agricolae" %in% rownames(installed.packages()) == 'FALSE') install.packages('agricolae')
if ("hms" %in% rownames(installed.packages()) == 'FALSE') install.packages('hms')1 Background
On July 5th, 6th, and 7th 2024 we conducted an ecotox assay on the development of Montipora capitata coral embryo development under exposure to PVC leachate at nominal concentrations of 0 mg/L (0.22um FSW, control), 0.01 mg/L (low), 0.1 mg/L (mid), and 1 mg/L (high). We repeated our assay across three nights of spawning. In the assay, embryos were incubated in 20mL scintillation vials in a climate-controlled lab. Here we detail the conditions of the incubation period.
We left a HOBO logger out on the lab bench, in the lab hood, and in an ambient flow through holding tank where adult coral colonies were being kept. We will compare the data here.
1.1 Inputs
- HOBO data logs
1.2 Outputs
- Visualizations comparing the lab bench conditions to the ambient seawater at HIMB around the July Montipora capitata spawning event in 2024.
2 Setup
2.1 Install packages
2.2 Load packages
library(tidyverse)
library(reshape2)
library(plotly)
library(car) # for Levene's test
library(agricolae) # for post-hoc tests
library(hms)2.3 Load HOBO logs
bench <- read.csv("embryo-inc-bench 2024-07-08 13_34_32 HST (Data HST).csv")
hood <- read.csv("embryo-inc-hood 2024-07-08 13_35_01 HST (Data HST).csv")
tank1 <- read.csv("Holding tank 2024-07-03 15_30_47 HST (Data HST).csv")
tank2 <- read.csv("Holding tank 2024-07-08 08_32_55 HST (Data HST).csv")
tank3 <- read.csv("AmbientTemp2 2024-07-08 08_32_20 HST (Data HST).csv")3 Format HOBO logs
bench <- bench %>%
rename(datetime = Date.Time..HST.,
temp = Temperature.....C.,
lux = Light....lux.) %>%
mutate(location = "bench") %>%
select(datetime, temp, lux, location)
hood <- hood %>%
rename(datetime = Date.Time..HST.,
temp = Temperature.....C.,
lux = Light....lux.) %>%
mutate(location = "hood") %>%
select(datetime, temp, lux, location)
tank1 <- tank1 %>%
rename(datetime = Date.Time..HST.,
temp = Temperature.....C.,
lux = Light....lux.) %>%
mutate(location = "tank1") %>%
select(datetime, temp, lux, location)
tank2 <- tank2 %>%
rename(datetime = Date.Time..HST.,
temp = Temperature.....C.,
lux = Light....lux.) %>%
mutate(location = "tank2") %>%
select(datetime, temp, lux, location)
tank3 <- tank3 %>%
rename(datetime = Date.Time..HST.,
temp = Temperature.....C.,
lux = Light....lux.) %>%
mutate(location = "tank3") %>%
select(datetime, temp, lux, location)all <- rbind(bench, hood, tank1, tank2, tank3)
all$datetime <- as.POSIXct(all$datetime, format = "%m/%d/%Y %H:%M:%S")4 Plot overlapping logs
long <- melt(all, id.vars=c("datetime", "location"), measure.vars=c("temp", "lux"))The incubation windows were:
1- July 5th 21:20 - July 6th 13:30
2 - July 6th 21:20 - July 7th 13:30
3- July 7th 21:20 - July 8th 13:30
incubation_windows <- data.frame(
start = as.POSIXct(c("07/05/2024 21:20:00",
"07/06/2024 21:20:00",
"07/07/2024 21:20:00"),
format = "%m/%d/%Y %H:%M:%S"),
end = as.POSIXct(c("07/06/2024 13:30:00",
"07/07/2024 13:30:00",
"07/08/2024 13:30:00"),
format = "%m/%d/%Y %H:%M:%S"))# Calculate scaling factor for the secondary (lux) axis
lux_scale <- max(all$temp, na.rm = TRUE) / max(all$lux, na.rm = TRUE)
ggplot(all, aes(x = datetime)) +
# Highlight windows
geom_rect(
data = incubation_windows,
aes(xmin = start, xmax = end),
ymin = -Inf, ymax = Inf, fill = "yellow", alpha = 0.2, inherit.aes = FALSE
) +
# Plot temp
geom_line(aes(y = temp, color = location), size = 1, alpha = 0.5) +
# Plot scaled lux
geom_line(aes(y = lux * lux_scale, color = location), linetype = "dashed") +
# Dual y-axis
scale_y_continuous(
name = "Temperature",
sec.axis = sec_axis(~ . / lux_scale, name = "Lux")
) +
theme_minimal() +
labs(color = "Location", title = "Temperature and Lux by Datetime with Incubation Highlights")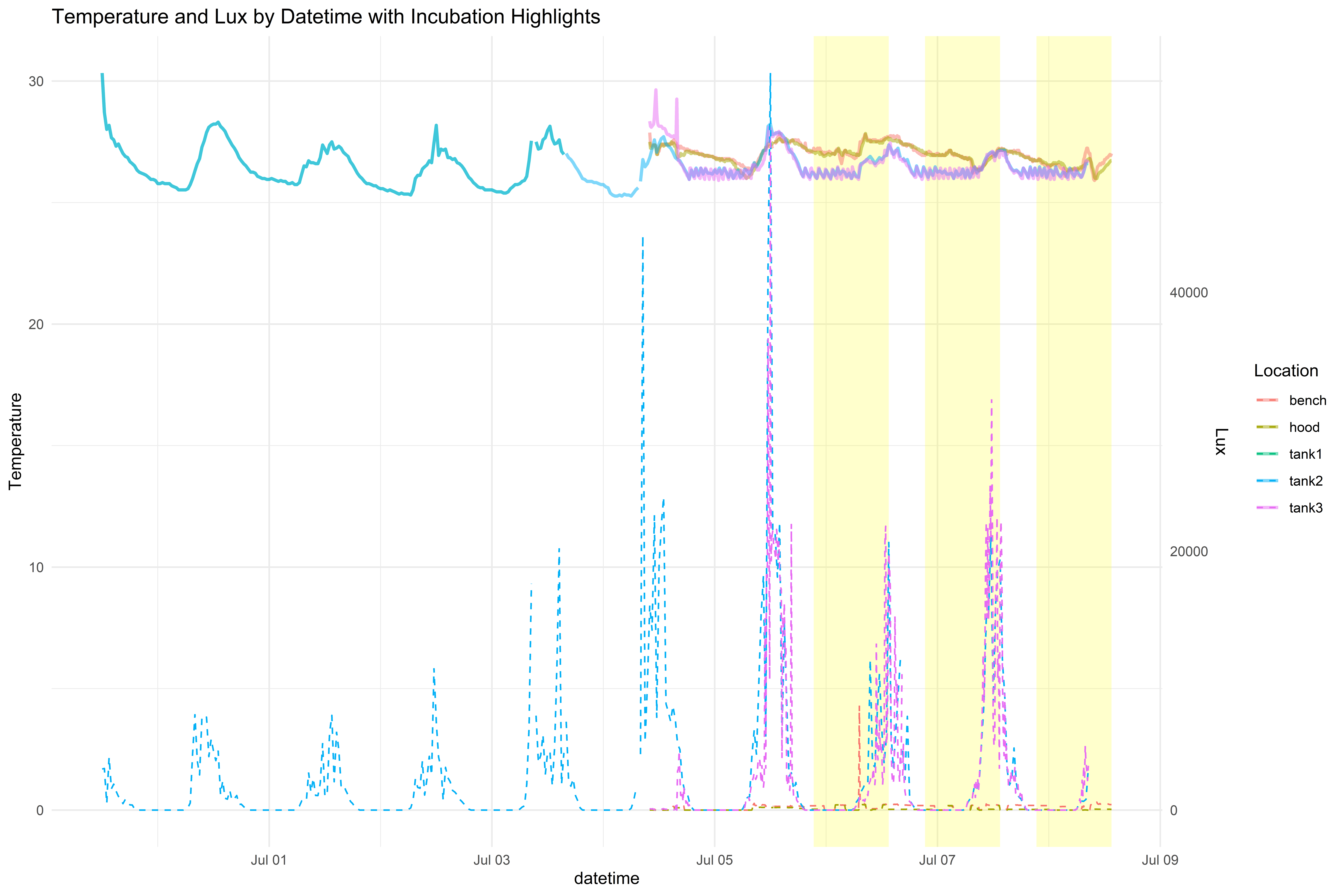
tank2 and tank3 look like they had heaters in them that kept the min temp up near 26F…
TO DO:
go back and find HOBO log or sea surface temp from July 5th - 8th 2024.
find calibration to convert lux to PAR
5 Averages
What were the average bench, hood, and tank temps & light measurements across the incubation periods?
Subset to the incubation windows
all_incubation <- all %>%
filter(
sapply(datetime, function(x) any(x >= incubation_windows$start & x <= incubation_windows$end))
)Display a table of averages and standard deviations
avg_sd <- all_incubation %>%
filter(location %in% c("bench", "hood" ,"tank2", "tank3")) %>%
group_by(location) %>%
summarize(
avg_temp = mean(temp, na.rm = TRUE),
sd_temp = sd(temp, na.rm = TRUE),
avg_lux = mean(lux, na.rm = TRUE),
sd_lux = sd(lux, na.rm = TRUE))
avg_sd| location | avg_temp | sd_temp | avg_lux | sd_lux |
|---|---|---|---|---|
| bench | 26.88575 | 0.3975264 | 243.58796 | 495.4855 |
| hood | 26.85602 | 0.3655191 | 91.64201 | 140.3716 |
| tank2 | 26.37685 | 0.3216703 | 2498.80348 | 5168.1143 |
| tank3 | 26.33780 | 0.3106606 | 2300.97458 | 5338.2473 |
Plot average temps
ggplot(avg_sd, aes(x = location, y = avg_temp, color = location)) +
geom_point(size = 3) + # mean temps as points
geom_errorbar(aes(ymin = avg_temp - sd_temp, ymax = avg_temp + sd_temp), width = 0.2) + # std dev error bars
labs(x = "Location", y = "Temperature (°C)", title = "Average Temperature with Standard Deviation") +
theme_minimal()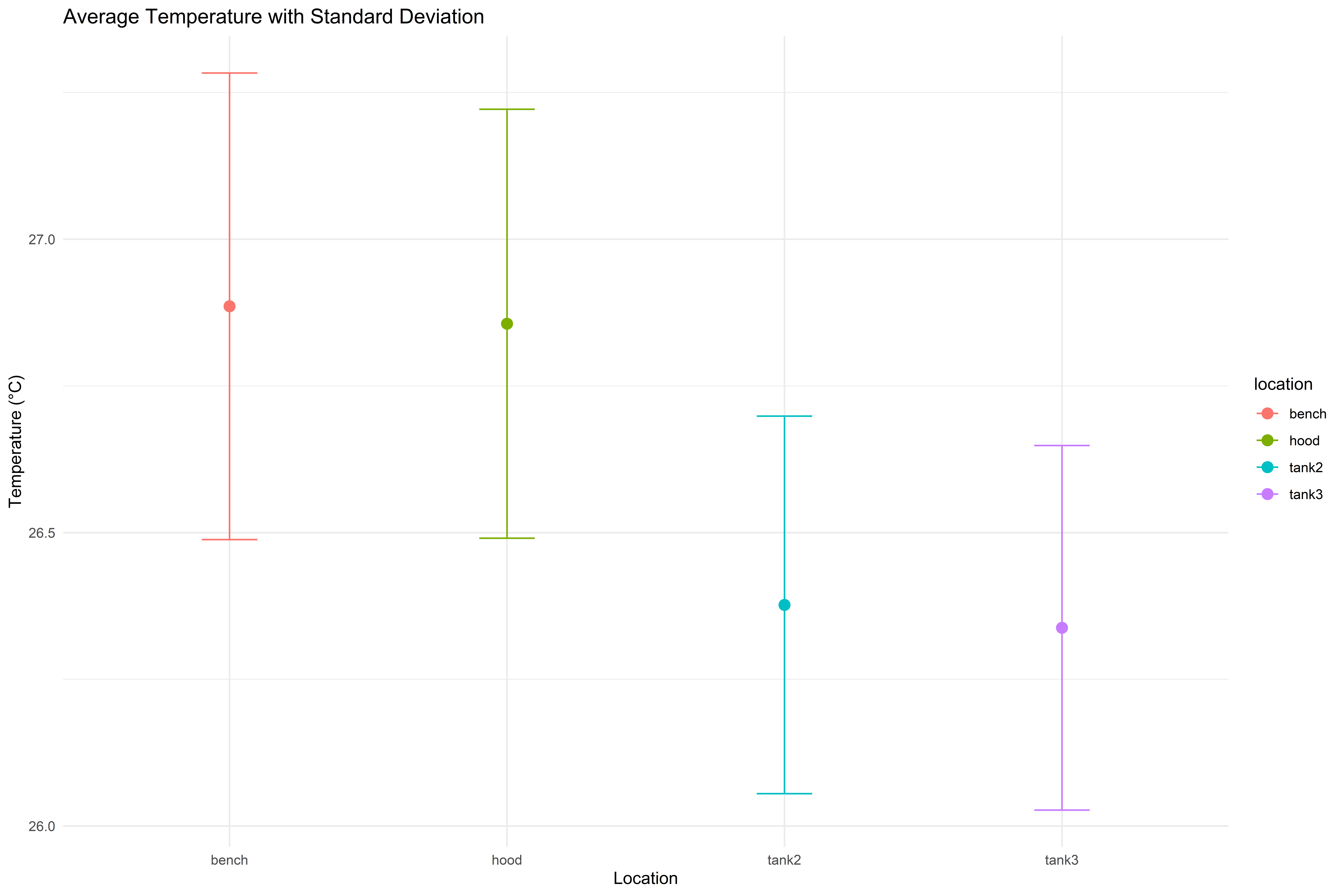
Plot average lux
ggplot(avg_sd, aes(x = location, y = avg_lux, color = location)) +
geom_point(size = 3) + # mean temps as points
geom_errorbar(aes(ymin = avg_lux - sd_lux, ymax = avg_lux + sd_lux), width = 0.2) + # std dev error bars
labs(x = "Location", y = "Lux", title = "Average lux with Standard Deviation") +
theme_minimal()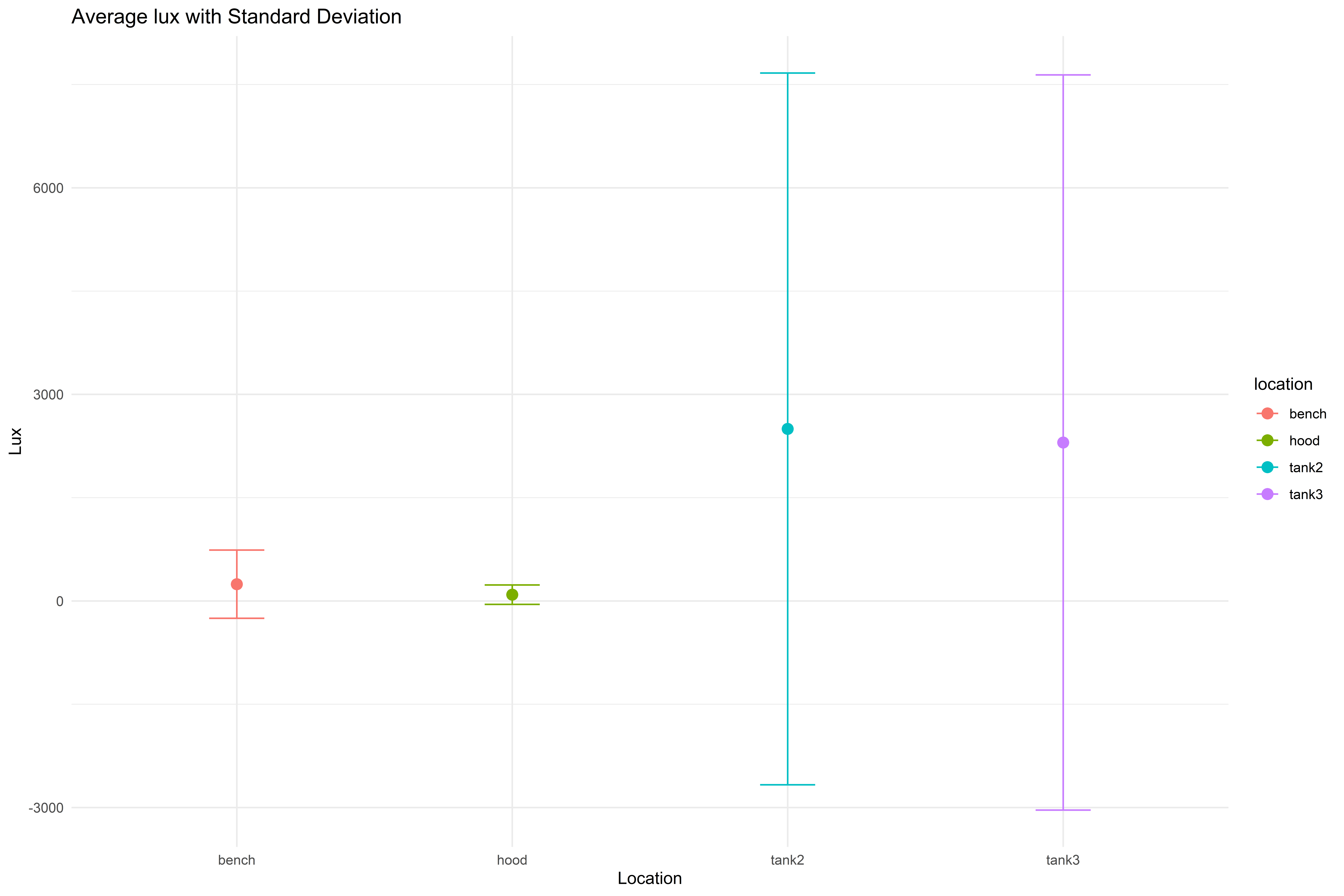
all_incubation <- all_incubation %>%
mutate(
space = case_when(
location %in% c("bench", "hood") ~ "lab",
location %in% c("tank2", "tank3") ~ "ambient",
TRUE ~ NA_character_ # for any other location values
)
)avg_sd <- all_incubation %>%
filter(space %in% c("lab", "ambient")) %>%
group_by(space) %>%
summarize(
avg_temp = mean(temp, na.rm = TRUE),
sd_temp = sd(temp, na.rm = TRUE),
min_temp = min(temp, na.rm = TRUE),
max_temp = max(temp, na.rm = TRUE),
avg_lux = mean(lux, na.rm = TRUE),
sd_lux = sd(lux, na.rm = TRUE),
min_lux = min (lux, na.rm=TRUE),
max_lux = max(lux, na.rm=TRUE))
avg_sd| space | avg_temp | sd_temp | min_temp | max_temp | avg_lux | sd_lux | min_lux | max_lux |
|---|---|---|---|---|---|---|---|---|
| ambient | 26.34765 | 0.3134719 | 25.95 | 27.37 | 2350.852 | 5289.2776 | 0 | 31733.76 |
| lab | 26.87088 | 0.3818228 | 25.91 | 27.84 | 167.615 | 371.6999 | 0 | 8069.12 |
The incubation temperature conditions were:
- ambient 26.34 +/- 0.31 C (min 25.95, max 27.37)
- lab 26.87 +/- 0.38 C (min 25.91, max 27.84)
The incubation lux conditions were:
- ambient 2350.85 +/- 5289.27 lux (min 0, max 31733.76)
- lab 167.61 +/- 371.69 lux (min 0, max 8069.12)
6 Hyopthesis testing
6.1 Is temp different across location?
# Check homogeneity of variance
leveneTest(temp ~ location, data = all_incubation)| Df | F value | Pr(>F) | |
|---|---|---|---|
| group | 3 | 4.863347 | 0.0023137 |
| 937 | NA | NA |
Our data has unequal variances (Levene’s test pvalue = 0.002314) so we use Welch’s t-test for ANOVA
# Welch's ANOVA comparing temperature by stage
welch_anova <- oneway.test(temp ~ location, data = all_incubation, var.equal = FALSE)
print(welch_anova)
One-way analysis of means (not assuming equal variances)
data: temp and location
F = 173.52, num df = 3.00, denom df = 351.78, p-value < 2.2e-16Follow up the Welch’s t-test with a post-hoc test
pairwise_results <- pairwise.t.test(
x = all_incubation$temp,
g = all_incubation$location,
p.adjust.method = "bonferroni",
pool.sd = FALSE
)
print(pairwise_results)
Pairwise comparisons using t tests with non-pooled SD
data: all_incubation$temp and all_incubation$location
bench hood tank2
hood 1 - -
tank2 <2e-16 <2e-16 -
tank3 <2e-16 <2e-16 1
P value adjustment method: bonferroni Pairwise comparisons of temperature across locations using Welch’s t-tests with Bonferroni-adjusted p-values revealed no significant difference between bench and hood (p = 1) or between tank2 and tank3 (p = 1). There is a significant difference (p<0.001) between the lab spaces (the bench and the hood) and the ambient spaces (The lab spaces are significantly different from the ambient spaces (tank2 and tank3).
6.2 Is temp different across space?
# Check homogeneity of variance
leveneTest(temp ~ space, data = all_incubation)| Df | F value | Pr(>F) | |
|---|---|---|---|
| group | 1 | 13.12683 | 0.0003067 |
| 939 | NA | NA |
Our data has unequal variances so we use Welch’s t-test for ANOVA
# Welch's ANOVA comparing temperature by stage
welch_anova <- oneway.test(temp ~ space, data = all_incubation, var.equal = FALSE)
print(welch_anova)
One-way analysis of means (not assuming equal variances)
data: temp and space
F = 520.18, num df = 1.00, denom df = 852.66, p-value < 2.2e-16Follow up the Welch’s t-test with a post-hoc test
pairwise_results <- pairwise.t.test(
x = all_incubation$temp,
g = all_incubation$space,
p.adjust.method = "bonferroni",
pool.sd = FALSE
)
print(pairwise_results)
Pairwise comparisons using t tests with non-pooled SD
data: all_incubation$temp and all_incubation$space
ambient
lab <2e-16
P value adjustment method: bonferroni 6.3 Is lab temp different across stage?
Add a column that indicates the duration of exposure by stage
all_incubation <- all_incubation %>%
mutate(
time_only = format(datetime, "%H:%M:%S"), # extract time as string
time_only = hms::as_hms(time_only), # convert to hms object for easier comparison
stage = case_when(
(time_only >= as_hms("21:20:00") | time_only <= as_hms("03:30:00")) ~ "cleavage", # crosses midnight
(time_only > as_hms("03:30:00") & time_only <= as_hms("08:30:00")) ~ "prawnchip", # normal range
(time_only > as_hms("08:30:00") & time_only <= as_hms("13:30:00")) ~ "early gastrula", # normal range
TRUE ~ NA_character_
)
)bench_by_stage <- all_incubation %>%
filter(!is.na(stage)) %>% # remove rows where stage is missing
filter(space == "lab") %>%
group_by(stage) %>%
summarize(
avg_temp = mean(temp, na.rm = TRUE),
sd_temp = sd(temp, na.rm = TRUE),
min_temp = min(temp, na.rm = TRUE),
max_temp = max(temp, na.rm = TRUE),
avg_lux = mean(lux, na.rm = TRUE),
sd_lux = sd(lux, na.rm = TRUE),
min_lux = min(lux, na.rm = TRUE),
max_lux = max(lux, na.rm = TRUE)
)
bench_by_stage| stage | avg_temp | sd_temp | min_temp | max_temp | avg_lux | sd_lux | min_lux | max_lux |
|---|---|---|---|---|---|---|---|---|
| cleavage | 26.87434 | 0.2328657 | 26.08 | 27.24 | 151.7127 | 161.3920 | 0.00 | 409.28 |
| early gastrula | 26.94533 | 0.5139978 | 25.91 | 27.71 | 201.0156 | 187.5445 | 0.81 | 613.60 |
| prawnchip | 26.79206 | 0.3650893 | 26.17 | 27.84 | 154.3572 | 619.0712 | 0.00 | 8069.12 |
The lab incubation temperature conditions were:
- cleavage 26.87 +/- 0.23 C (min 26.08, max 27.24)
- prawnchip 26.79 +/- 0.36 C (min 26.17, max 27.84)
- early gastrula 26.94 +/- 0.51 C (min 25.91, max 27.71)
The lab incubation lux conditions were:
- cleavage 151.7127 +/- 161.39 lux (min 0, max 409.28)
- prawnchip 154.35 +/- 619.07 lux (min 0, max 8069.12)
- early gastrula 201.01 +/- 187.54 (min 0.81, max 613.60)
lab_incubation <- all_incubation %>%
filter(!is.na(stage)) %>% # remove rows where stage is missing
filter(space == "lab")
# Check homogeneity of variance
leveneTest(temp ~ stage, data = lab_incubation)| Df | F value | Pr(>F) | |
|---|---|---|---|
| group | 2 | 73.8332 | 0 |
| 585 | NA | NA |
Our data has unequal variances so we use Welch’s t-test for ANOVA
# Welch's ANOVA comparing temperature by stage
welch_anova <- oneway.test(temp ~ stage, data = lab_incubation, var.equal = FALSE)
print(welch_anova)
One-way analysis of means (not assuming equal variances)
data: temp and stage
F = 5.9606, num df = 2.00, denom df = 320.16, p-value = 0.002873Follow up the Welch’s t-test with a post-hoc test
pairwise_results <- pairwise.t.test(
x = lab_incubation$temp,
g = lab_incubation$stage,
p.adjust.method = "bonferroni",
pool.sd = FALSE
)
print(pairwise_results)
Pairwise comparisons using t tests with non-pooled SD
data: lab_incubation$temp and lab_incubation$stage
cleavage early gastrula
early gastrula 0.2608 -
prawnchip 0.0269 0.0037
P value adjustment method: bonferroni Pairwise comparisons of temperature across stage using Welch’s t-tests with Bonferroni-adjusted p-values revealed the temperatures experienced during the latest embryonic stage (early gastrula) are significantly different from the prawnchip stage (p<0.05). HOWEVER, the actual temperature range experienced across embryonic development (26.79-26.94) is within the ambient seawater range and is ecologically parallel to the natural sea surface temp fluctuations experienced in a diurnal cycle.
6.4 Is lab temp different across spawn night?
Add a column that indicates the spawn night (1, 2, or 3)
lab_incubation <- lab_incubation %>%
mutate(
spawn = case_when(
datetime >= incubation_windows$start[1] & datetime <= incubation_windows$end[1] ~ 1,
datetime >= incubation_windows$start[2] & datetime <= incubation_windows$end[2] ~ 2,
datetime >= incubation_windows$start[3] & datetime <= incubation_windows$end[3] ~ 3,
TRUE ~ NA_integer_
)
)Print a table of temps and lux by spawn night
by_spawn <- lab_incubation %>%
group_by(spawn) %>%
summarize(
avg_temp = mean(temp, na.rm = TRUE),
sd_temp = sd(temp, na.rm = TRUE),
min_temp = min(temp, na.rm = TRUE),
max_temp = max(temp, na.rm = TRUE),
avg_lux = mean(lux, na.rm = TRUE),
sd_lux = sd(lux, na.rm = TRUE),
min_lux = min(lux, na.rm = TRUE),
max_lux = max(lux, na.rm = TRUE)
)
by_spawn| spawn | avg_temp | sd_temp | min_temp | max_temp | avg_lux | sd_lux | min_lux | max_lux |
|---|---|---|---|---|---|---|---|---|
| 1 | 27.23163 | 0.2814402 | 26.60 | 27.84 | 213.6418 | 594.5645 | 0 | 8069.12 |
| 2 | 26.85444 | 0.2351807 | 26.21 | 27.24 | 149.9682 | 167.1667 | 0 | 472.48 |
| 3 | 26.52658 | 0.2326195 | 25.91 | 27.24 | 139.2350 | 176.6241 | 0 | 613.60 |
lab_incubation <- lab_incubation %>%
mutate(spawn = factor(spawn, levels = c(1, 2, 3),
labels = c("1", "2", "3"))) # convert spawn to factor with labels
# Check homogeneity of variance
leveneTest(temp ~ spawn, data = lab_incubation)| Df | F value | Pr(>F) | |
|---|---|---|---|
| group | 2 | 10.11787 | 4.79e-05 |
| 585 | NA | NA |
Our data has unequal variances so we use Welch’s t-test for ANOVA
# Welch's ANOVA comparing temperature by stage
welch_anova <- oneway.test(temp ~ spawn, data = lab_incubation, var.equal = FALSE)
print(welch_anova)
One-way analysis of means (not assuming equal variances)
data: temp and spawn
F = 366.72, num df = 2.00, denom df = 387.43, p-value < 2.2e-16Follow up the Welch’s t-test with a post-hoc test
pairwise_results <- pairwise.t.test(
x = lab_incubation$temp,
g = lab_incubation$spawn,
p.adjust.method = "bonferroni",
pool.sd = FALSE
)
print(pairwise_results)
Pairwise comparisons using t tests with non-pooled SD
data: lab_incubation$temp and lab_incubation$spawn
1 2
2 <2e-16 -
3 <2e-16 <2e-16
P value adjustment method: bonferroni Pairwise comparisons of temperature across spawn night using Welch’s t-tests with Bonferroni-adjusted p-values revealed that each night is significantly different (p<2.2e-16). HOWEVER the actual range of temps across all three spawn nights is 26.5 to 27.2 (less than 1C), and is an ecological parallel to actual fluctuations in sea surface temp that occur across nights of spawning.
7 Summary
Temperature Although statistically significant differences in temperature were detected across both spawn nights (with temperatures generally decreasing over the three nights) and developmental stage (with the lowest temperatures observed just before dawn during the prawnchip stage, but no significant difference between the cleavage and early gastrula stages), the magnitude of these differences was small and remained within the normal range of ambient seawater temperature fluctuations observed during a spawning event. Therefore, while temperature varied somewhat across the experimental assay, this variation is minor and unlikely to confound our results, as embryos in nature develop under similar thermal conditions.
Light